Bioinformatics is a field of science that combines biology and computer science to analyze biological data. Bioinformatics tools help organize, compare, and analyze data, predict protein structures, conduct genomic analysis, and support research.
In this article, I have compiled some tools that will be beneficial not only for bioinformaticians but also for researchers, scientists, analysts, students, and more.
I will discuss various tools that can assist you in analyzing biological data, performing statistical analysis, visualizing data, calling variants, predicting protein structures, using genome browsers, conducting sequence alignment, and more.
Best Bioinformatics Tools

OmicsBox
OmicsBox stands out as a top-notch bioinformatics software, providing thorough analysis of Next-Generation Sequencing (NGS) data for genomes, transcriptomes, and metagenomes. It’s widely used by leading research institutions globally, designed to be user-friendly, efficient, and packed with powerful tools for handling big and complex datasets.
OmicsBox is structured in modules, each tailored for specific analyses, including genome analysis, genetic variation, transcriptomics, functional analysis, and metagenomics.

Key Features
- The Genome Analysis Module is an efficient and user-friendly toolset to characterize and analyze newly sequenced genomes.
- The Transcriptomics Module allows for processing RNA-Seq data from raw reads to functional analysis flexibly and intuitively.
- The Functional Analysis Module provides biological context as an analysis option.
- The Metagenomics Module enables microbiome data analysis, including assembly, annotation, and classification of metagenomic data.
OmicsBox provides a suite of tools for analyzing genomic, transcriptomic, and metagenomic data, making it valuable for professionals working in computational biology and bioinformatics.
Bioconductor
Bioconductor, an open-source effort, includes specialized R packages for interpreting high-throughput genomic data. R language and environment for statistical computing and graphics is enhanced by Bioconductor to address the unique challenges posed by genomics and bioinformatics.
The comprehensive toolkit provided by Bioconductor allows users to perform a range of tasks, from basic data preprocessing to advanced statistical analyses.
Key Features
- Bioconductor toolkit provide a vast array of specialized R packages for tasks such as the analysis of microarray data, high-throughput sequencing data, flow cytometry data, and more.
- High-quality documentation and reproducible research
- Bioconductor packages often include functionalities to interface with and retrieve data from popular genomic databases, such as Ensembl and UCSC Genome Browser.
Bioconductor toolkit is widely used by bioinformaticians, researchers, and scientists who work with high-throughput genomic data.
FastQC
FastQC is a widely used quality control tool designed for sequence data generated through next-generation sequencing methods. FastQC offers a straightforward approach to quality checks on raw data from sequencing pipelines.
The tool is easily downloadable and features a user-friendly interface, allowing users to address data issues before proceeding with further analysis. Input files consist of read sequences, and the tool generates output in the form of graphics and tabular summaries of the results.
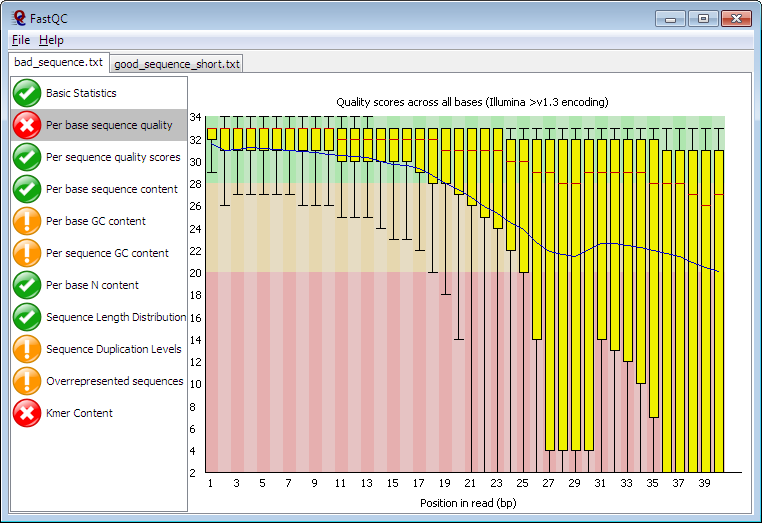
Kay Features
- Reports can be automatically generated without using the interactive application when offline.
- Providing a quick overview of the problem.
- Import data from BAM, SAM, or FastQ files (any variant) for convenient analysis.
- Generates summary graphs and tables for data quality evaluation
- HTML Report Export
It is Ideal for processing and analyzing sequencing data. So, It is Recommended for biological data analysts, researchers, and project teams.
EMBOSS
EMBOSS (European Molecular Biology Open Software Suite) is one of the tools tools to learn bioinformatical analysis if you are a beginner. EMBOSS is a cutting-edge, free, and open-source software package meticulously crafted to meet the unique demands of the molecular biology and bioinformatics community.
EMBOSS has many tools for analyzing sequences, and it works well with other popular packages and tools. One of them is EMBOSS Needle:
Key Features
EMBOSS includes over 200 applications for:
- Showing features of a sequence
- Database searching
- Presentation of sequence data
- 3D molecular models
- Bioinformatics platforms
- Splitting a sequence into smaller sequences
It is a beginner-friendly tool useful for students, computational researchers, bioinformaticians, and data scientists.
Clustal
Clustal is a set of bioinformatics tools such as ClustalX, ClustalW, and Clustal Omega. It is designed for aligning biological sequences like DNA, RNA, and proteins. They help identify similar regions in multiple sequences, aiding in the analysis of evolutionary relationships and functional domains.
The Clustal series of programs is widely used in molecular biology for the multiple alignment of both nucleic acid and protein sequences and for preparing phylogenetic trees.
Key Features
- Clustal software typically offers user-friendly interfaces
- Clustal excels at aligning three or more sequences
- Clustal X helps predict multiple sequence alignment and phylogenetic analysis for given gene sequences of various organisms.
- Clustal Omega uses a heuristic based on phylogenetic analysis
Clustal is best suited for professionals in the fields of bioinformatics and computational biology who need to align and analyze multiple biological sequences.
DNASTAR Lasergene
DNASTAR Lasergene is a leading-edge software suite that serves as a versatile solution for molecular biology, genomics, and protein analysis. This comprehensive platform is relied upon by scientists worldwide.
Additionally, DNASTAR offers Nova Applications, including NovaFold AI, NovaFold, NovaFold Antibody, and NovaDock, further enhancing its capabilities in protein structure prediction and analysis.
Key Features
- Comprehensive solutions for molecular biology, antibody, genomics, and protein analysis, offering a complete suite for researchers.
- Automates tasks in genomics projects, streamlining sequence assembly and analysis.
- Provides flexible and rich 3D graphical representations of protein structures, enhancing the understanding of protein sequences.
DNASTAR Lasergene, with its comprehensive packages and advanced Nova Applications, is a versatile software suite that caters to the diverse needs of molecular biologists, genomics researchers, protein scientists, and structural bioinformaticians.
GATK
The Genome Analysis Toolkit (GATK), created by the Broad Institute’s Data Science platform, is a powerful tool for discovering genetic variants and genotyping. It excels in processing large input files and focuses on identifying variations like SNPs and indels in DNA and RNA-Seq data.
GATK also handles copy numbers and structural variations and includes utilities for quality control in high-throughput sequencing.
Key Features
- GATK is optimized for accurate and high-quality results.
- GATK employs a structured programming framework, leveraging the functional programming philosophy of MapReduce.
- It maximizes computational efficiency for efficient processing.
- Capable of genomic analysis for exomes and whole genomes.
- Offers best-practice workflows for somatic short variants.
GATK is tool for those aiming to navigate the complexities of genomic analysis with accuracy and ease such as genomic researchers, scientists, and bioinformaticians.
Universal Analysis Software
Universal Analysis Software (UAS) simplifies the analysis and management of forensic genomic data. It supports various ForenSeq workflows, rapidly processes data, and provides reliable variant calls without requiring per-seat licenses.
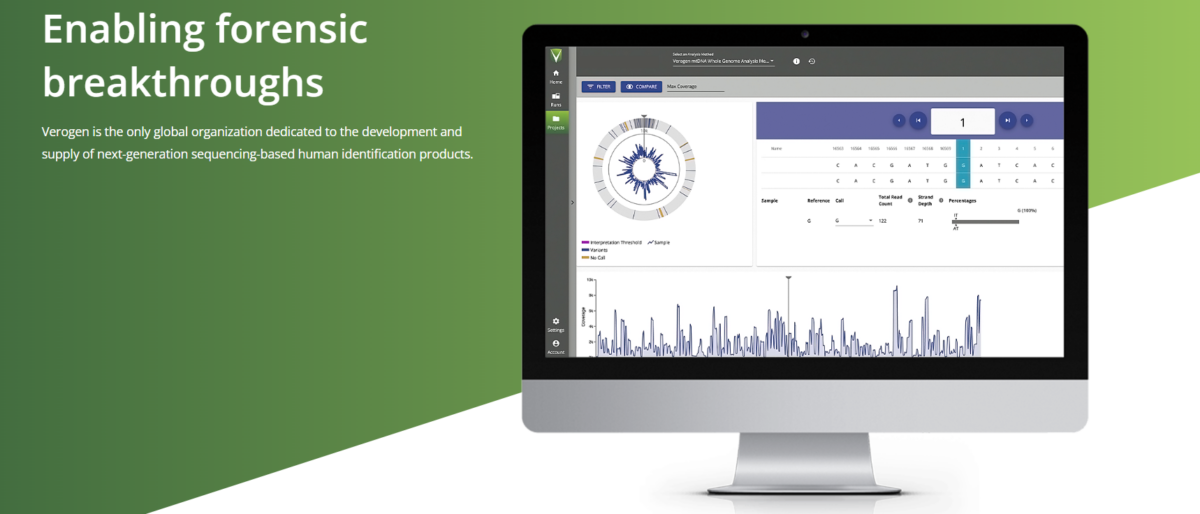
Tailored for forensic analysts, UAS streamlines the handling of sequence information, facilitating efficient review of DNA profiles. UAS is designed for compatibility with the MiSeq FGx Sequencing System and common third-party tools, making it a user-friendly and versatile solution.
Key Features
- Secure interface with MiSeq FGx System for automated post-sequencing data analysis.
- Intuitive tools for sample management, run setup, data visualization, and reporting.
- Real-time run monitoring, sample comparisons, and intensity plots for informed decision-making.
- Preinstalled on a dedicated server for powerful computing without infrastructure hassles.
Universal Analysis Software is a specialized tool that simplifies forensic genomic data analysis, offering a range of features for forensic analysts and laboratories.
TinyBio
Tinybio is a pioneering genomic generative AI company that simplifies processes for real scientists with user-friendly tools and software. It focuses on enhancing productivity and resource optimization, allowing scientists to concentrate on their research without software complexities.
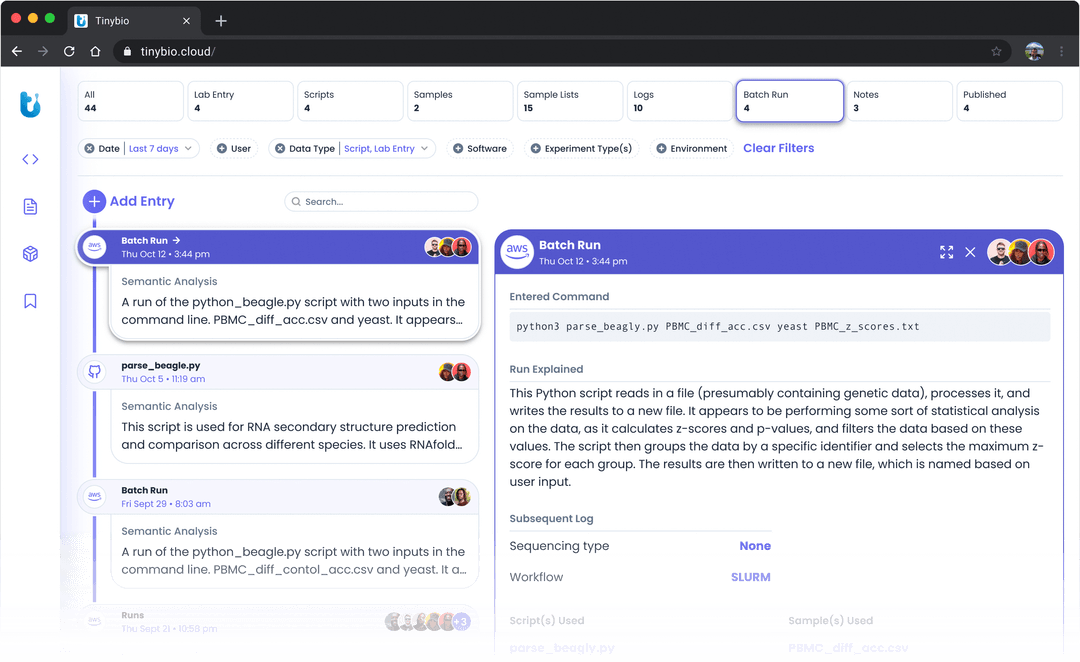
Utilizes an AI tool to understand and suggest possible analyses on a set of samples. Tinybio is best suited for those in genomics research who seek AI-driven solutions to streamline their workflows, from generating experiments to constructing analysis pipelines.
Key Features
- Features a chatbot designed to ideate, debug, and navigate complexities in bioinformatics.
- Specialized tool for rapidly constructing biology analysis pipelines.
- A specialized tool to reverse engineer the code responsible for producing a scientific paper.
- Provides a solution to organize the computing environment without migration.
- User-Centric Approach
The platform’s user-centric approach and comprehensive tool suite make it a valuable asset for bioinformaticians, scientists, computational biologists, and researchers in both academia and industry.
DeepVariant
DeepVariant, a deep learning technology, was developed through collaboration between the Google Brain team and Verily Life Sciences. This innovative tool is designed to tackle the challenge of reconstructing precise and comprehensive genome sequences from high-throughput sequencing (HTS) data.
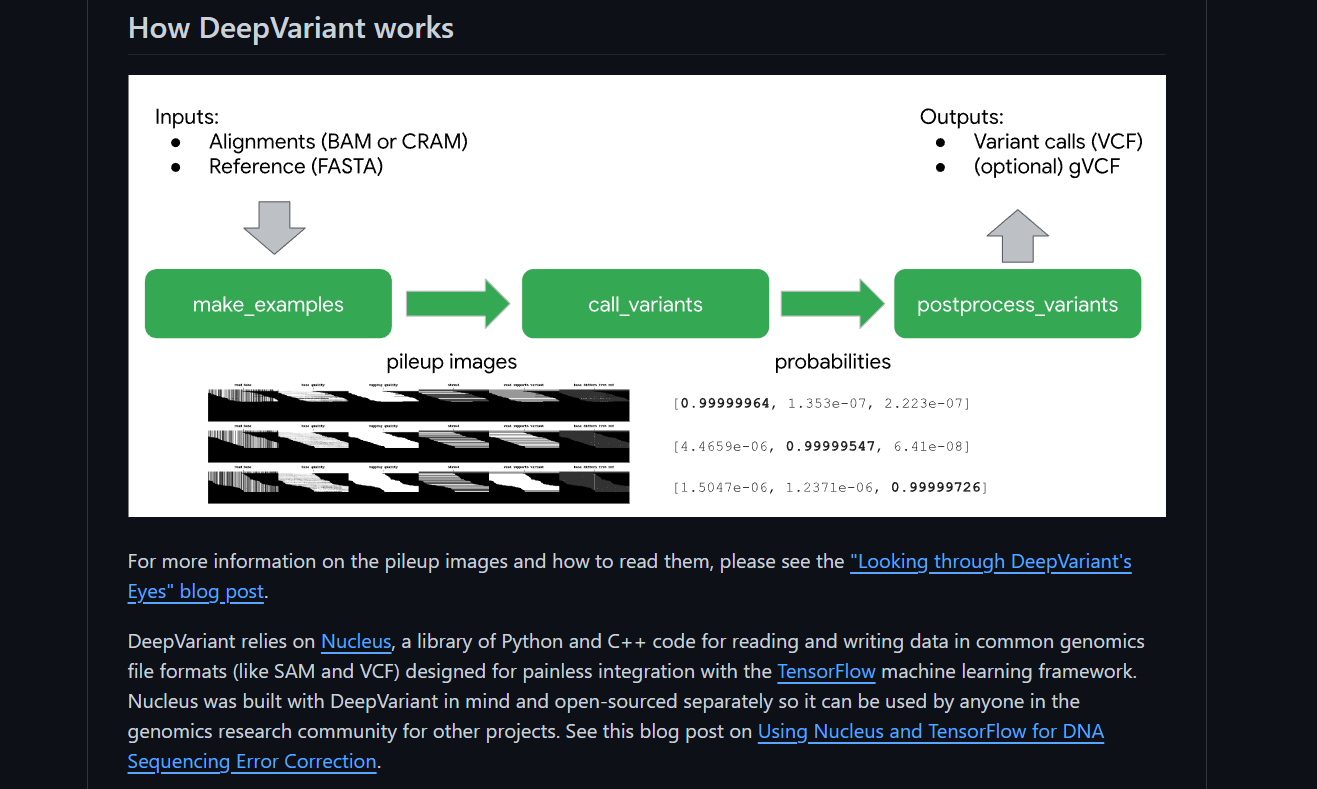
HTS generates around 1 billion short DNA sequences (reads), representing only a fraction of the entire genome. It is an open-source analysis pipeline that uses a deep neural network to call genetic variants from next-generation DNA sequencing data.
Key Features
- DeepVariant uniquely applies deep learning techniques to address the challenges of variant calling, offering improved accuracy compared to traditional methods.
- The transformation of the variant calling problem into an image classification problem sets DeepVariant apart.
- Partnered with GCP to deploy DeepVariant workflows on the cloud
DeepVariant is a helpful tool for genomic research, using deep learning and being open-source for researchers and bioinformaticians.
Galaxy
Galaxy is an open-source, web-based platform for data-intensive biomedical research. It’s used by scientists to analyze large biomedical datasets like those in genomics, proteomics, metabolomics, and imaging.
With a graphical interface, it supports various biological data formats and is open-source. Its applications include gene expression, proteomics, transcriptomics, and more.
Key Features
- It provides thousands of tools to choose from
- User-Friendly Interface and Intuitive Graphical Interface
- Galaxy supports a variety of biological data formats
- Provides CPU and disk space which is enough to analyze large datasets
The galaxy will be helpful for Bioinformaticians, drug researchers, and computational biologists who can use it since applied overboard to the field of Bioinformatics.
DNAnexus
DNAnexus, in genomics by developing an API-based platform dedicated to facilitating the sharing and management of data and tools essential for accelerating genomic research.
DNAnexus provides a global network for genomics, allowing scientists and clinicians worldwide to collaborate securely on genomic research related to various fields, including cancer, heart disease, Alzheimer’s disease, noninvasive prenatal testing, and agriculture.
Key Features
- DNAnexus’s products include Data management solutions, Data analysis solutions, Collaboration solutions, Biomedical data analysis, and Software Bioinformatics applications.
- The platform operates on an API-based infrastructure, simplifying data and tool management and accelerating genomic research processes.
- DNAnexus addresses challenges in genome informatics and data portability by providing a cloud-based solution.
DNAnexus provides a secure, scalable, and compliant platform for bioinformaticians, enterprises, and researchers globally.
Autodock
AutoDock is a suite of automated docking tools designed to predict how small molecules, such as drug candidates, bind to a receptor with a known 3D structure.
AutoDock is best for applications in X-ray crystallography, structure-based drug design, lead optimization, virtual screening, combinatorial library design, protein-protein docking, and chemical mechanism studies.

Key Features
- AutoDock facilitates the docking of ligands (small molecules) to a set of grids representing the target protein.
- AutoDock 4 comprises auto dock for ligand docking and autogrid for pre-calculating grids describing the target protein.
- AutoDockTools (ADT) is a graphical user interface facilitating the setup of rotatable bonds in the ligand and the analysis of docking.
- AutoDock Vina internally calculates grids for atom types needed for docking, eliminating the need for users to choose atom types and pre-calculate grid maps.
AutoDock’s versatility makes it a valuable tool for a broad range of scientific applications, particularly in the life sciences and drug development sectors. It is helpful for researchers and biologists.
Rosetta
The Rosetta software suite encompasses algorithms for the computational modeling and analysis of protein structures. Its application has led to significant breakthroughs in computational biology, facilitating accomplishments such as de novo protein design, enzyme design, ligand docking, and the structure prediction of biological macromolecules and complexes.

Key Features
- Understanding macromolecular interactions
- Designing custom molecules
- Creating effective methods for exploring conformation and sequence space.
- Finding broadly useful energy functions for various biomolecular representations
Rosetta’s toolkit is helpful for scientists, computational biologists, students, and researchers engaged in macromolecular modeling and structural biology.
BioJava
BioJava serves as a specialized bioinformatic platform designed for the Java environment, catering to the processing needs of diverse biological data.
The platform executes various operations such as sequence manipulation, protein structure analysis, Distributed Annotation System (DAS) utilization, and dynamic programming and supports interoperability with Common Object Request Broker Architecture (CORBA).
Key Features
- BioJava’s capabilities include managing local PDB installations, manipulating structures, conducting standard analyses like aligning sequences and structures, and 3D visualization.
- It facilitates data retrieval from databases for nucleotide and protein sequences.
- BioJava also supports tasks like reading and writing sequence file formats, translating DNA sequences to proteins, and executing common bioinformatics routines.
- It enables searching for similar sequences and the manipulation of individual sequences.
BioJava is well-suited for individuals and researchers in the field of bioinformatics, scientists, and computational biologists who prefer utilizing Java-based tools for processing diverse biological data.
Bionano
Bionano’s solutions offer unparalleled resolution across all classes of genomic variation, addressing the limitations of traditional tools and methods. The Bionano Analysis Software (Variant Intelligence Applications) integrates OGM data with NGS, chromosomal microarray (CMA), and other data types, enabling comprehensive interpretation and analysis of variant data. Bionano’s focus areas span clinical care, research, and therapeutics.

Key Features
- VIA Analysis Software uses microarray and NGS data to assess HRD
- Simplifies genome assembly, structural variant analysis, and hybrid scaffolding
- Saphyr System detects and analyzes structural variants with high speed and throughput
- Nexus Copy Number enables visual and statistical analysis of genetic variation in research cohorts.
It is an invaluable tool for genomic researchers, clinicians, and biotechnologists aiming to push the boundaries of genomic understanding and application.
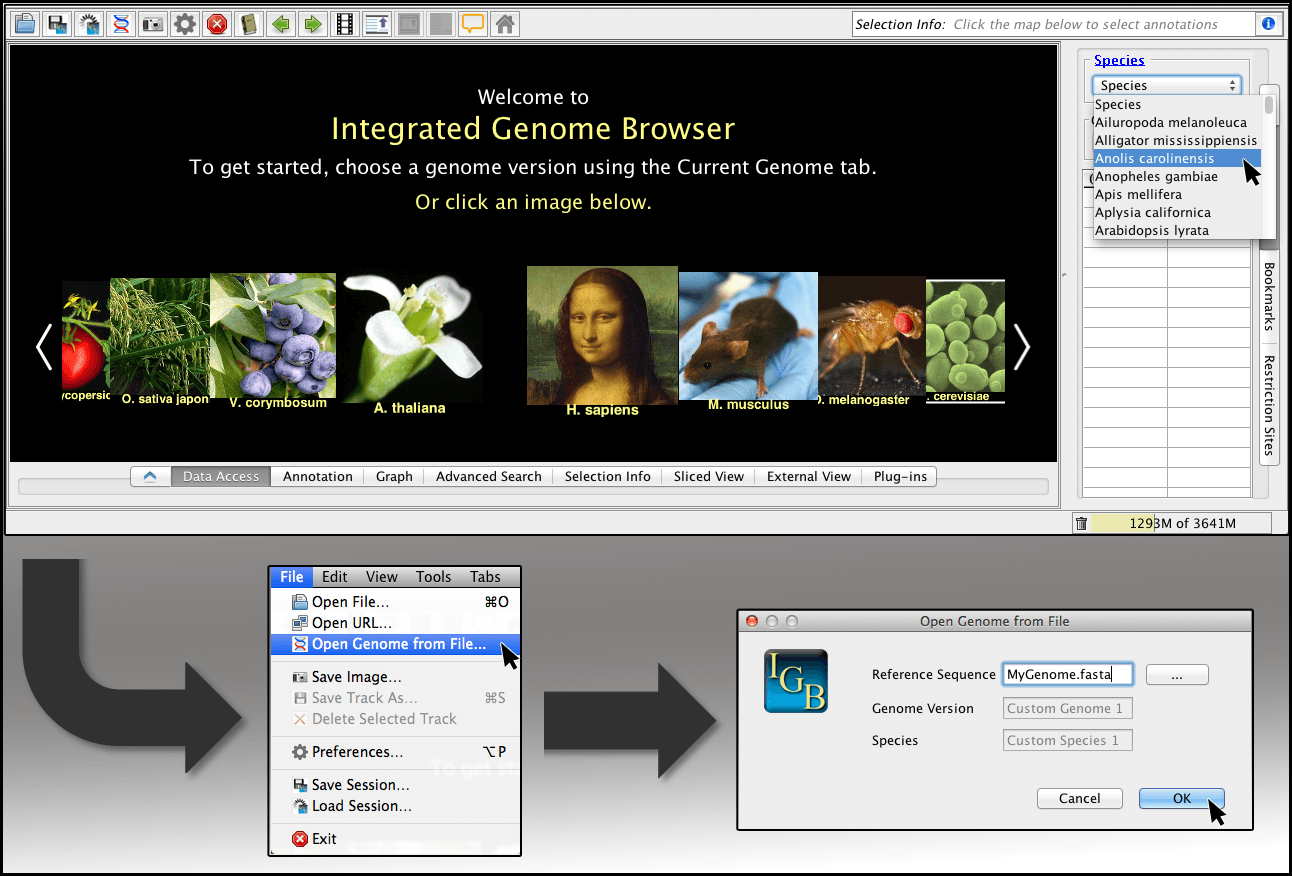
Integrated Genome Browser
The Integrated Genome Browser serves as a visualization tool designed to illustrate intricate biological patterns within genomics datasets, sequence data, gene models, and DNA microarray data.
Compatible with UNIX, Linux, Mac, and Windows operating systems, this software offers a reliable and swift solution for visualizing extensive data directly on a desktop.
Key Features
- Streamline your workflow by employing scripts to define a genome. You can also utilize R for managing IGB.
- Make, customize, and save graphs from your data. Use depth graphs to show coverage or mismatch graphs to count differences between your data and a reference.
- You can also make your plug-ins/apps and share them with the IGB community.
- You can build an IGB QuickLoad to share data between collaborators. Store your data in the cloud to provide access to everyone involved in the project.
Integrated Genome Browser is a helpful tool for data experts, scientists, and bioinformaticians to automate workflow.
DRAGEN
DRAGEN analysis finds applications across various fields in the biological sciences, offering a transformative approach to genomic analysis. The DRAGEN analysis provides a precise, thorough, and streamlined secondary examination of next-generation sequencing data.
DRAGEN analysis, leveraging highly reconfigurable FPGA technology, accelerates genomic analysis algorithms while addressing challenges in computing times and data volumes.
Key Features
- Analyzes complete genomes, exomes, methylomes, and transcriptomes using a unified platform.
- Utilizes graph reference genome and machine learning, driving unprecedented accuracy.
- Identify and profile infectious diseases with an all-encompassing solution.
- DRAGEN analysis provides the flexibility to insert a variety of input files and produce a range of output documents.
DRAGEN analysis proves to be highly beneficial to genomic analysis. It is helpful for computational biologists, and genomic analysts.
PathAI
PathAI leverages computer vision and deep learning to analyze pathology images, transforming the traditional approach to cancer diagnosis and treatment.
The three main applications of PathAI’s products include aiding pathologists in making better diagnoses, enhancing drug development through informed therapeutic use, and extending gold-standard pathology services to regions lacking access.

Key Features
- Automates monotonous tasks for pathologists, enhancing efficiency in analyzing pathology slides.
- Seamlessly integrates histopathology with genomic data, providing valuable insights to inform therapeutic decisions.
- Uses a variety of data and outcomes, along with multiple scanners, stains, and lab sources, to make predictions better while avoiding bias.
By combining pathologists’ expertise with PathAI’s software, the platform frees up time and enables a focus on determining optimal treatment options for individual patients.
Geneious
Geneious software, developed in Java Swing, boasts a high level of interoperability across commonly used operating systems. It serves as a comprehensive biological analysis tool with a user-friendly interface.
Supports multiple functionalities such as multiple alignments, phylogenetic trees, contig assemblies, statistical graphs, 3D structures, chromatograms, and electropherograms.

Key Features
- Next-Generation Sequencing Assembly and Analysis
- Visualizations and Graphics
- Geneious enables the retrieval of biological databases from various platforms.
- Editing and Assembly of Chromatogram
- Alignment and Phylogenetics
Geneious is a widely known Bioinformatics software, helpful for researchers, bioinformaticians, and computational biologists.
MEGA
MEGA, short for Molecular Evolutionary Genetics Analysis, is a comprehensive software package crafted for the exploration and visualization of molecular sequence data. Its primary focus lies in investigating the evolutionary relationships embedded within DNA or protein sequences.
MEGA equips researchers with versatile tools, enabling them to conduct phylogenetic analyses, estimate evolutionary distances, and construct detailed phylogenetic trees.
Key Features
- MEGA allows users to construct phylogenetic trees to visualize and analyze the evolutionary relationships among biological sequences.
- Various statistical approaches are integrated into MEGA for assessing the significance of results in molecular evolution analyses.
- MEGA provides visualization tools for exploring and interpreting complex molecular data, including sequence alignments and phylogenetic trees.
Researchers, bioinformaticians, and scientists use MEGA to gain insights into the evolutionary history of genes, proteins, and other biological entities.
BLAST
The Basic Local Alignment Search Tool (BLAST) identifies local similarities between sequences by comparing nucleotide or protein sequences to databases. It calculates the statistical significance of matches, allowing for the inference of functional and evolutionary relationships between sequences and aiding in the identification of gene family members.
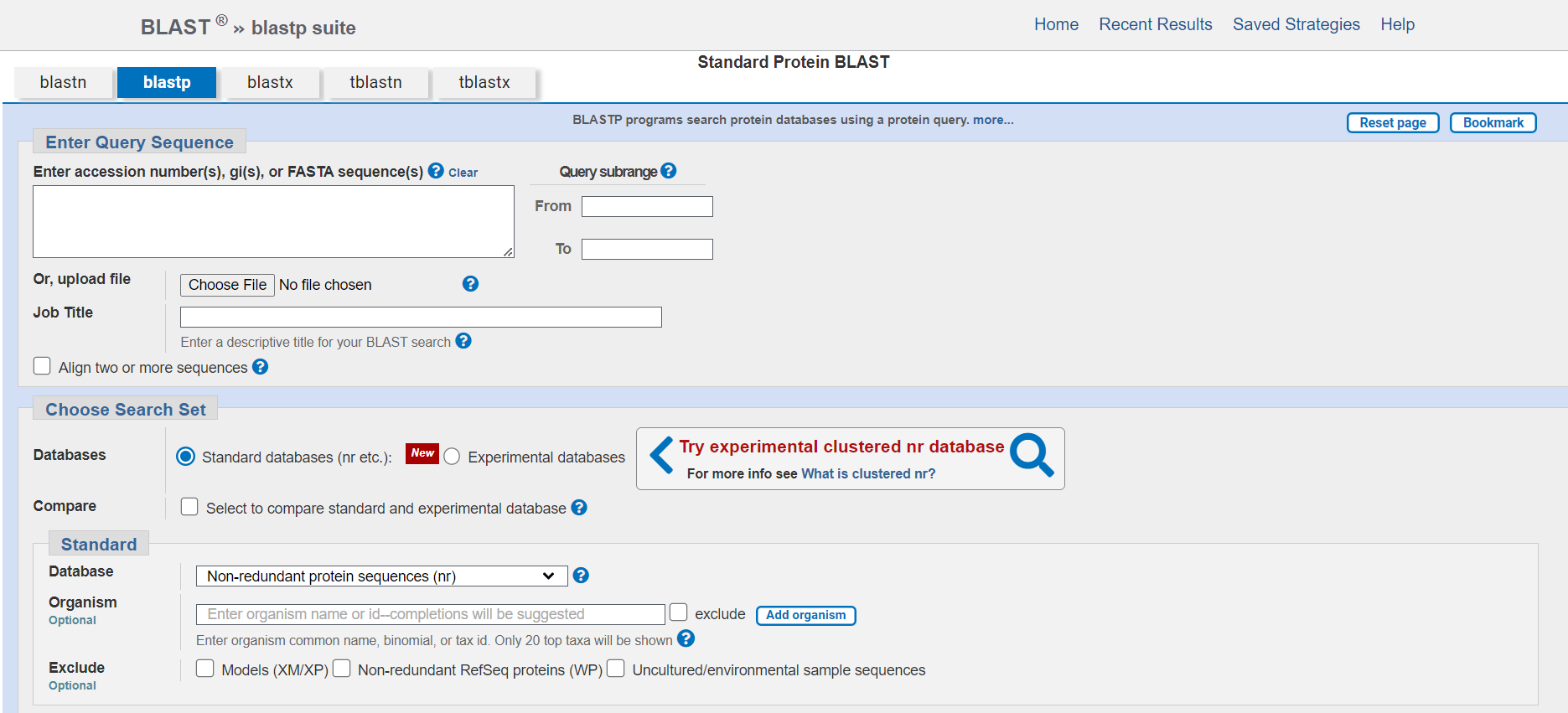
BLAST is a widely used bioinformatics program and algorithm for comparing primary biological sequence information, such as the amino-acid sequences of proteins or the nucleotides of DNA sequences.
Key Features
- BLAST is primarily used for comparing a query sequence against a database of sequences to find similar or homologous sequences.
- There are several versions of BLAST, including nucleotide BLAST (for nucleotide sequences), Protein BLAST (for protein sequences), BLASTx (translates nucleotide sequences to protein sequences before comparing), and others.
- BLAST can search against various biological databases, including the NCBI nucleotide and protein databases.
BLAST is a vital tool in bioinformatics and molecular biology, essential for swiftly and accurately identifying similarities in DNA, RNA, and protein sequences.
Clara Parabricks
Clara Parabricks is a cost-effective software suite for the rapid secondary analysis of NGS DNA and RNA data, delivering exceptionally fast results compared to other methods. As a comprehensive genomic analysis tool, Parabricks significantly enhances throughput time for tasks like germline and somatic analysis.
For example, it can analyze 30x whole human genome data in just 25 minutes, a remarkable improvement over the 30 hours typically required. Its output aligns with widely used software, ensuring easy result verification.
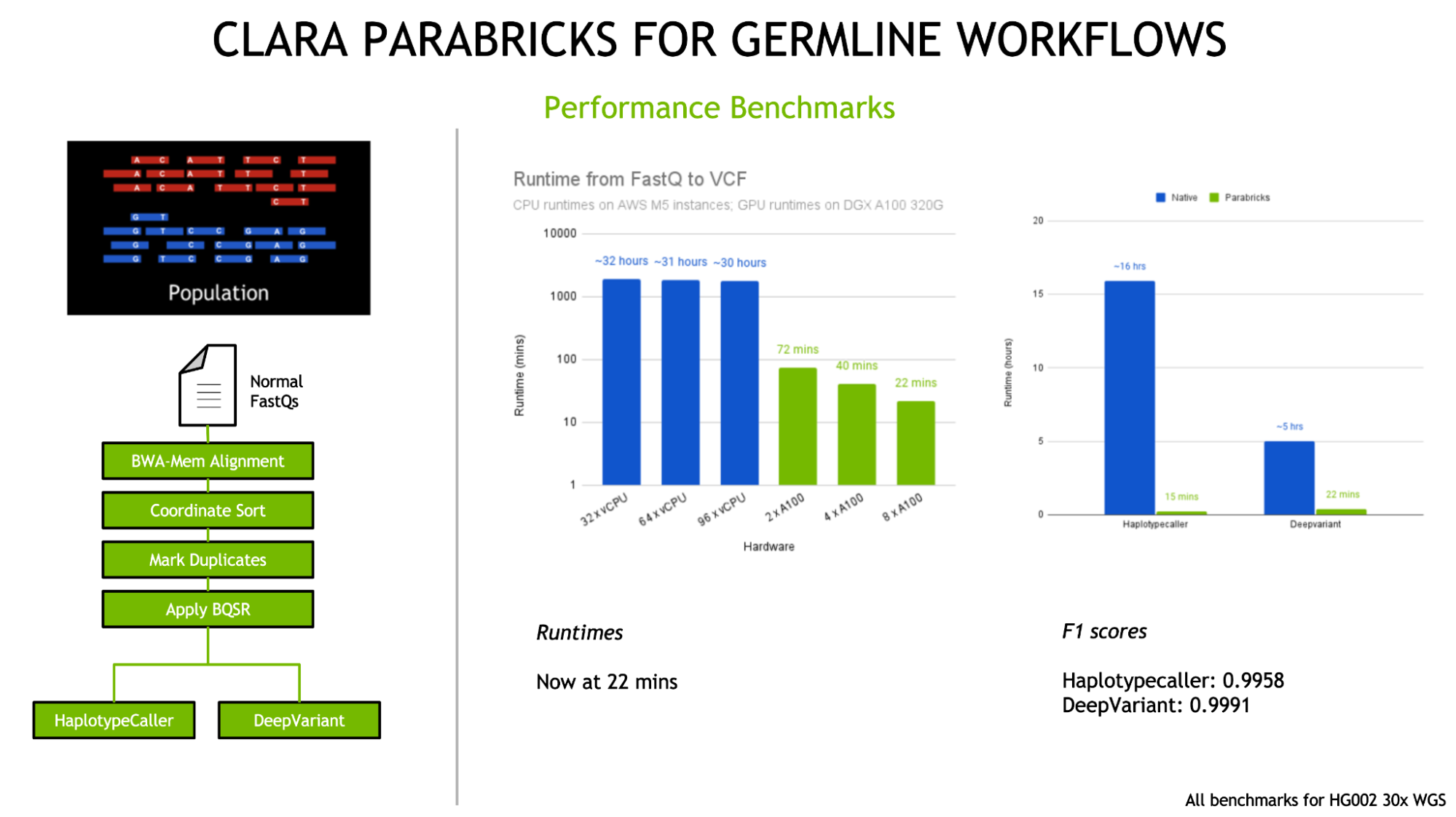
Key Features
- Clara Parabricks is uniquely GPU-accelerated, optimizing secondary analysis with remarkable speed
- Integration of deep learning-based tools alongside industry-standard ones.
- Significantly reduced compute time and analysis costs.
- Compatibility with Azure and free availability on NGC.
- Versatility in supporting various genomics workflows.
Clara Parabricks is ideal for genomic researchers, bioinformaticians, clinical labs, pharmaceutical companies, educational institutions, and healthcare professionals.
Pluto
Pluto is a versatile and user-friendly platform revolutionizing scientific research. It simplifies project management, allowing teams to organize, monitor, and assign experiments with secure data sharing for efficient collaboration.
Scientists benefit from its robust data analysis and visualization tools, supporting computational biology algorithms for insights into gene expression, DNA-protein binding, and biomarkers.
Key Features
- Empowerment of scientists to run powerful bioinformatics analyses directly in the browser.
- Transformation of raw data into publication-ready plots in minutes.
- Elegant platform for organizing, monitoring, and assigning experiments.
- Intuitive tools for analyzing and visualizing complex biological data.
- Compatibility with various biological assays, including RNA-seq, ChIP-seq, CUT&RUN, and plate-based assays.
Pluto is a versatile platform designed for users in biological research, computational biology, project management, and collaborative scientific efforts. With its user-friendly interface and comprehensive features, it holds value for diverse roles within the scientific research community.
LatchBio
Latchbio is the most flexible platform to access and analyze data for biological R&D. It is a biological cloud where organizations can store, process, analyze, and visualize multi-omics data.
You can upload bioinformatics workflows in any language using the Latch SDK, receive associated no-code interfaces for your scientists, and benefit from a highly scalable infrastructure that supports your entire company.

Key Features
- Latch Registry is a user-friendly database with a spreadsheet interface designed to facilitate intricate metadata capture for Next-Generation Sequencing (NGS) and multi-omics files within the Latch platform.
- Latch Data is a versatile file storage system capable of hosting limitless data and providing universal access for all members of your organization through a unified login.
- Latch Pods stand out as agile and robust cloud computing units, featuring pre-installed RStudio and JupyterLab for effortless downstream analysis of workflow results.
Latchbio allows you to access and analyze your data with a user-friendly interface. Hence, it is a helpful tool for bioinformaticians, wet lab scientists, and team leads.
Final Words
Bioinformatics tools are essential for understanding and unraveling the complexities of biological data. From organizing information to predicting protein structures and conducting genomic analysis, these tools benefit researchers, scientists, and students alike.
As technology gets better, teamwork between biology and computer science keeps helping us learn more about life. The tools mentioned in this article will assist you in analyzing biological data, conducting statistical analysis, visualizing data, calling variants, predicting protein structures, utilizing genome browsers, performing sequence alignment, and more.


